ARTÍCULO DE REVISIÓN
REVISTA DE LA FACULTAD DE MEDICINA HUMANA 2021 - Universidad Ricardo PalmaDOI 10.25176/RFMH.v21i1.3327
MOLECULAR BASIS OF COVID-19 PATHOGENESIS AND IN SILICO STUDIES OF POTENTIAL PHARMACOLOGICAL TREATMENT
BASES MOLECULARES DE LA PATOGÉNESIS DE COVID-19 Y ESTUDIOS IN SILICO DE POSIBLES TRATAMIENTOS FARMACOLÓGICOS
Eduardo Lam-Cabanillas(1,a), Anthony León-Risco(1,a), Kevyn León-Risco(1,a), Gaby Llamo-Hoyos(1,a), Rosa López-Zavaleta(1,a), Estefanía Luzuriaga-Tirado(1,a), Alex Mendoza-Blas(1,a), Jorge Huamán-Saavedra(1,b)
1 Facultad de Medicina, Universidad Nacional de Trujillo. Trujillo, Peru.
a Medical Student.
b Medical Doctor.
COVID-19 is a disease caused by SARS-CoV-2, a virus that represents a serious threat to global health. The objective of the article is to deepen into the structure of the non-structural proteins (Nsp1-16) and structural proteins (Spike, Envelope, Mmebrane and Nucleocapside) of SARS-CoV-2 and their role during the infection of its target cells; also to propose molecules from in silico studies that inhibit proteins of the viral cycle for potential pharmacological treatments. The SARS-CoV-2 genome has ORF1a and ORF1ab that encode sixteen non-structural proteins (Nsp1-16), as well as thirteen ORFs that encode four structural proteins and nine accessory proteins. The Nsps participate both in the viral cycle, being Nsp3 and Nsp5 responsible for the cleavage of polyproteins 1a and 1ab for the subsequent formation of the replica-transcriptase complex, and favoring the progress of the viral infection. The S protein mediates the union and fusion of the virus to the host cell through its subunits S1 and S2, respectively; however, it must be previously activated by proteases such as TMPRSS2, furin, trypsin or catepsins. On the other hand, the E, M and N proteins participate in the assembly of the virus and until now the functions of the accessory proteins are unknown. Also, in silico studies of drugs such as disulfiram and viomycin, and molecules found in plants as Azadirachta indica, the tea plant, and Andrographis paniculata have shown inhibitory effects on the SARS-CoV-2 viral cycle.
Keywords: COVID-19, SARS-CoV-2, genome, protein structure, in silico. (Source: MeSH – NLM)
RESUMEN
La COVID-19 es una enfermedad causada por el virus SARS-CoV-2, que representa una grave amenaza para la salud mundial. El objetivo de este artículo es profundizar en la estructura de las proteínas no estructurales (Nsp1-16) y estructurales (Spike, Envoltura, Membrana y Nucleocápside) del SARS-CoV-2, su rol durante la infección a sus células diana; asimismo plantear moléculas de estudios in silico que inhiben a las proteínas del ciclo viral para posibles tratamientos farmacológicos. El genoma del SARS-CoV-2 presenta los ORF1a y ORF1ab que codifican dieciséis proteínas no estructurales (Nsps), así como trece ORFs que codifican cuatro proteínas estructurales y nueve accesorias. Las Nsps participan tanto en el ciclo viral, siendo Nsp3 y Nsp5 responsables del clivaje de las poliproteinas 1a y 1ab para la posterior formación del complejo replicasa-transcriptasa, como favoreciendo el progreso de la infección viral. La proteína S media la unión y fusión del virus a la célula huésped a través de sus subunidades S1 y S2, respectivamente; sin embargo, se debe activar previamente por proteasas como TMPRSS2, furina, tripsina o catepsinas. Por otra parte, las proteínas E, M y N participan en el ensamblaje del virus y hasta el momento se desconoce las funciones de las proteínas accesorias. Asimismo, estudios in silico de medicamentos como el disulfiram y el viomycin, y moléculas de la Azadirachta indica, la planta de té y la Andrographis paniculata han mostrado efectos inhibitorios en el ciclo viral del SARS-CoV-2.
Palabras Claves: COVID-19, genoma, in silico. (Fuente: DeCs - BIREME)
COVID-19 is a disease caused by the SARS-CoV-2 virus, a new betacoronavirus that represents a serious threat to global health (1). The first cases of COVID-19 were reported in late December 2019 in Wuhan-China, and three months later it had already spread around the world (1); for this reason, the World Health Organization (WHO) declared it a pandemic on March 11th of this year(1). Since its emergence to the date, the number of infected people worldwide has exceeded 36 754 395 confirmed cases and 1 064 838 deaths (10/10/2020)(2). As a part of these figures, Peru has more than 843 355 confirmed cases and 33 158 deaths to date (10/10/2020)(3), making it one of the countries with the highest number of deaths in South America and the world.
On January 24th, 2020, Zhu N. et al(4) published the genetic sequence of SARS-CoV-2 as a result of a real-time reverse transcription PCR clinical trial from patients with atypical pneumonia in Wuhan City. Thanks to this discovery it was determined that SARS CoV-2 has a 5' untranslated region (UTR), a replicase complex (ORF1ab), S, E, M, N genes, a 3'-UTR and other open reading frames (ORFs) that differ from the rest of betacoronavirus(4,5).
Despite being a new virus, the study of the genome and proteins of coronaviruses with which it shares a large percentage of identity has been developed for several years, research mainly conducted in SARS-CoV and MERS-CoV, which has provided the basis for the study of SARS-CoV-2 proteins(6,7).
Due to the importance of certain proteins in replication, transcription and assembly processes of SARS-CoV-2, multiple studies have been carried out with drugs for its inhibition, as treatment of COVID-19(8); among them are hydroxychloroquine, azithromycin and remdesivir that have been studied for a long time without any favorable results(8,9). However, several researchers have focused their efforts on experimenting with other molecules using in silico studies, as they are faster and cheaper, for later in vivo or in vitro studies.
The main goal of this investigation is to deepen in the molecular aspect the pathogenesis of COVID-19, through the description of the role of the non-structural proteins (Nsps) and structural proteins of SARS-CoV-2(4) in the infection of its target cells. Also, to show possible pharmacological treatments of in silico studies that have as purpose the inhibition of essential proteins in the development of the viral cycle and could serve as reference for later in vitro and in vivo studies.
The procedure to search for information was initiated by searching for all non-structural proteins (Nsps) and structural proteins of SARS-CoV-2 in MeSH. Once they were identified, the search of each one of them was made until September 6th in PubMed, Scielo, Google Scholar and LinkSpringer databases, using as criteria publication in indexed journals, which respond to the purpose of this publication and have relevant information. In this search, 100 articles were found and 68 were selected, most of them from PubMed; only 4 had the exception of being preprint due to the relevance of their information.
To create Figure 2 and Figure 3,
the identification codes of each protein were obtained from the Protein Data Bank(10) (PDB), then its amino acidic sequence was visualized with the program Jalview(11) and using its extension Quimera(12) the 3D structure of each protein was visualized. The program CellPAINT(13) was used to draw the virus, the cell and the structural proteins of SARS-CoV-2 in Figure 4 as indicated by the authors(13). The program BioRender(14) was also used, using its pre-designed figures of the cell and organelles, adding the structure of the virus and its proteins to schematize the pathogenesis process of SARS-CoV-2 in Figure 5(14).
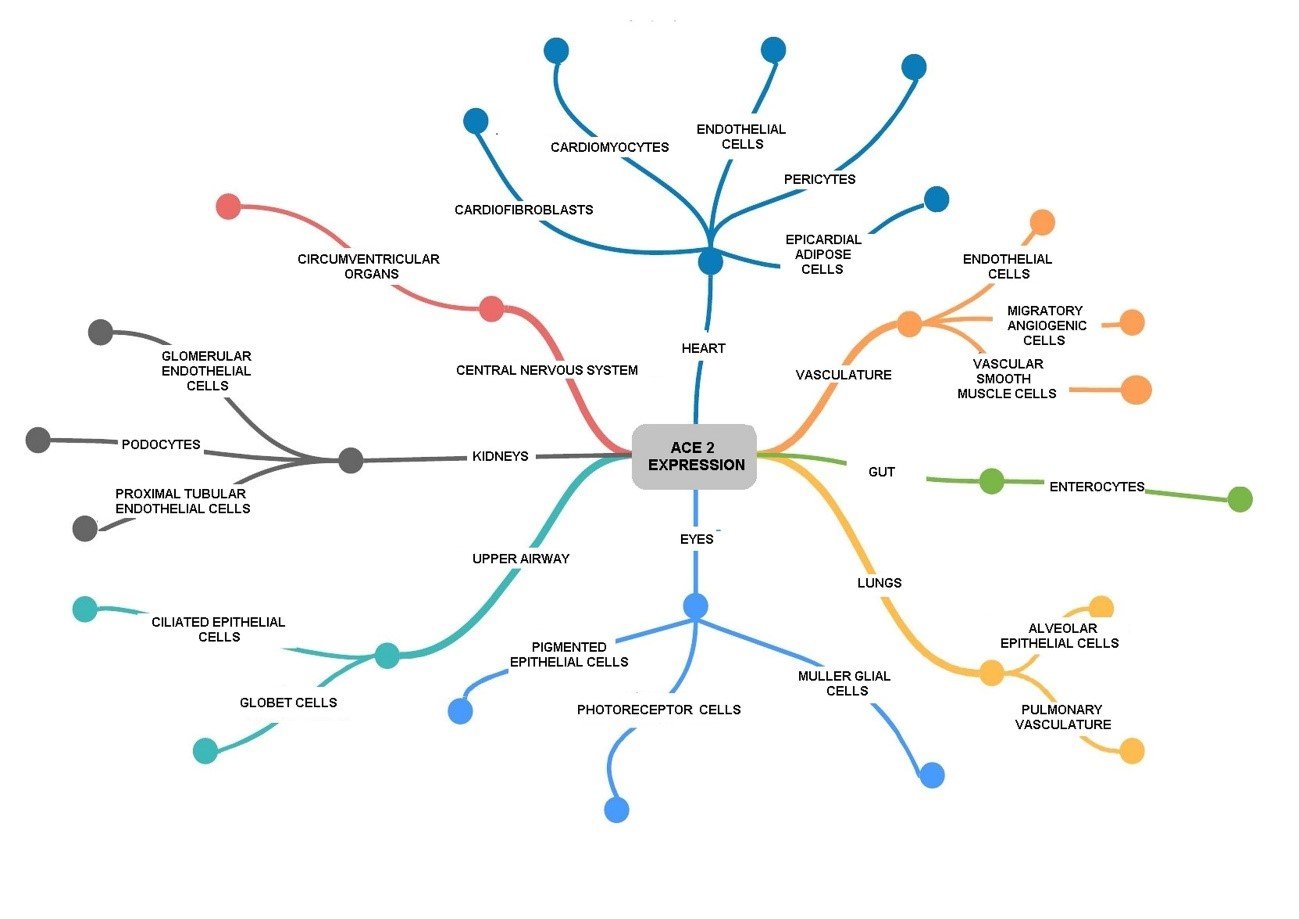
Figure 2. Three-dimensional protease structure of SARS-CoV-2. PLpro (papain-like protease) domain of Nsp3 (PDB: 6W9C) and Nsp5 (PDB: 6Y2E) with its 3CLpro (3-chymotrypsin-like protease) domain. This figure was made using UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.
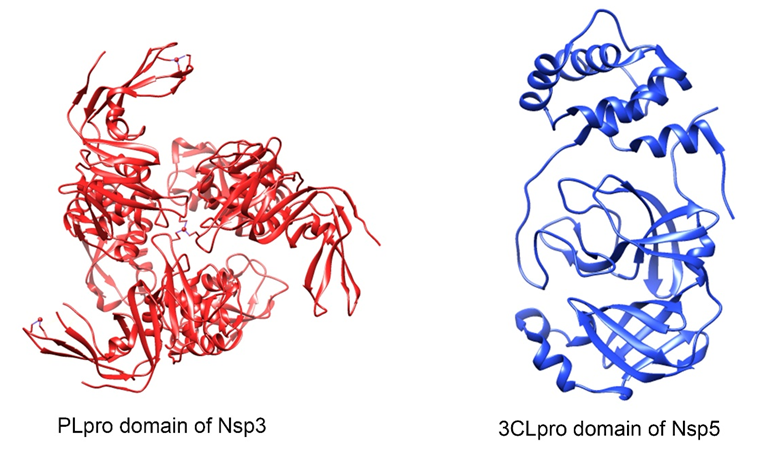
Figure 3. Three-dimensional protein structure of the SARS-CoV-2 replicase-transcriptase complex. Nsp7-Nsp8 (turquoise Nsp7, magenta Nsp8) (PDB: 7JLT), Nsp10 (PDB: 6W75), Nsp12 (PDB: 7BW4), Nsp13 (PDB: 6ZSL) and Nsp16 (PDB: 6W75) heterodimers. This figure was made using UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.
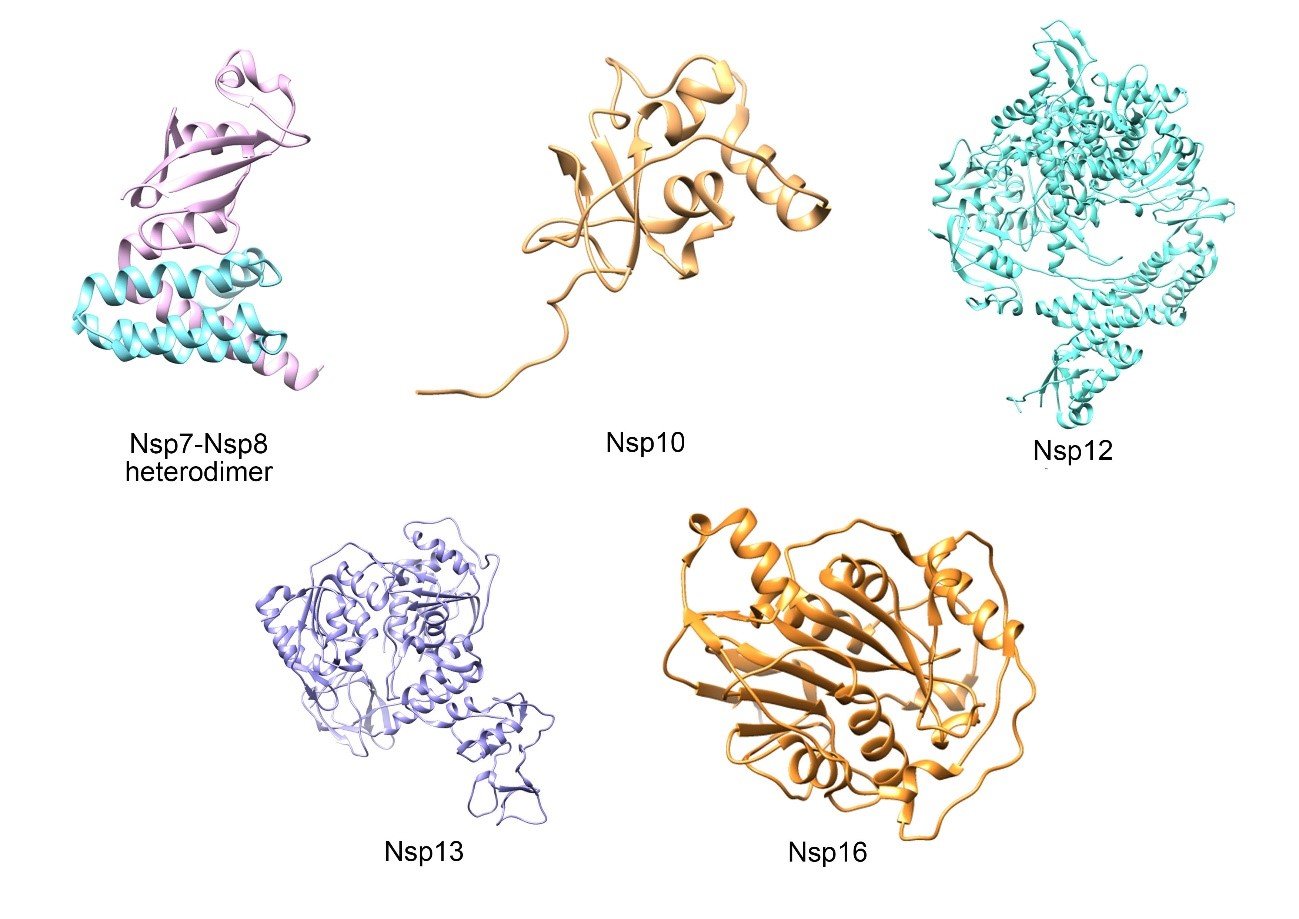
Figure 4. Binding of SARS-CoV-2 to ACE2. Structural proteins Spike (S) pink, Membrane (M) turquoise, Envelope (E) blue and Nucleocapsid (N) red. Three-dimensional structure of the Spike protein (PDB: 6VXX) and its binding to ECA2 (yellow) in the host cell membrane. This figure was made using Gardner A, Autin L, Barbaro BA, Olson AJ, Goodsell DS. CellPAINT: Interactive Illustration of Dynamic Mesoscale Cellular Environments. IEEE Comput Graph Appl. 2018;38(6):51-66.
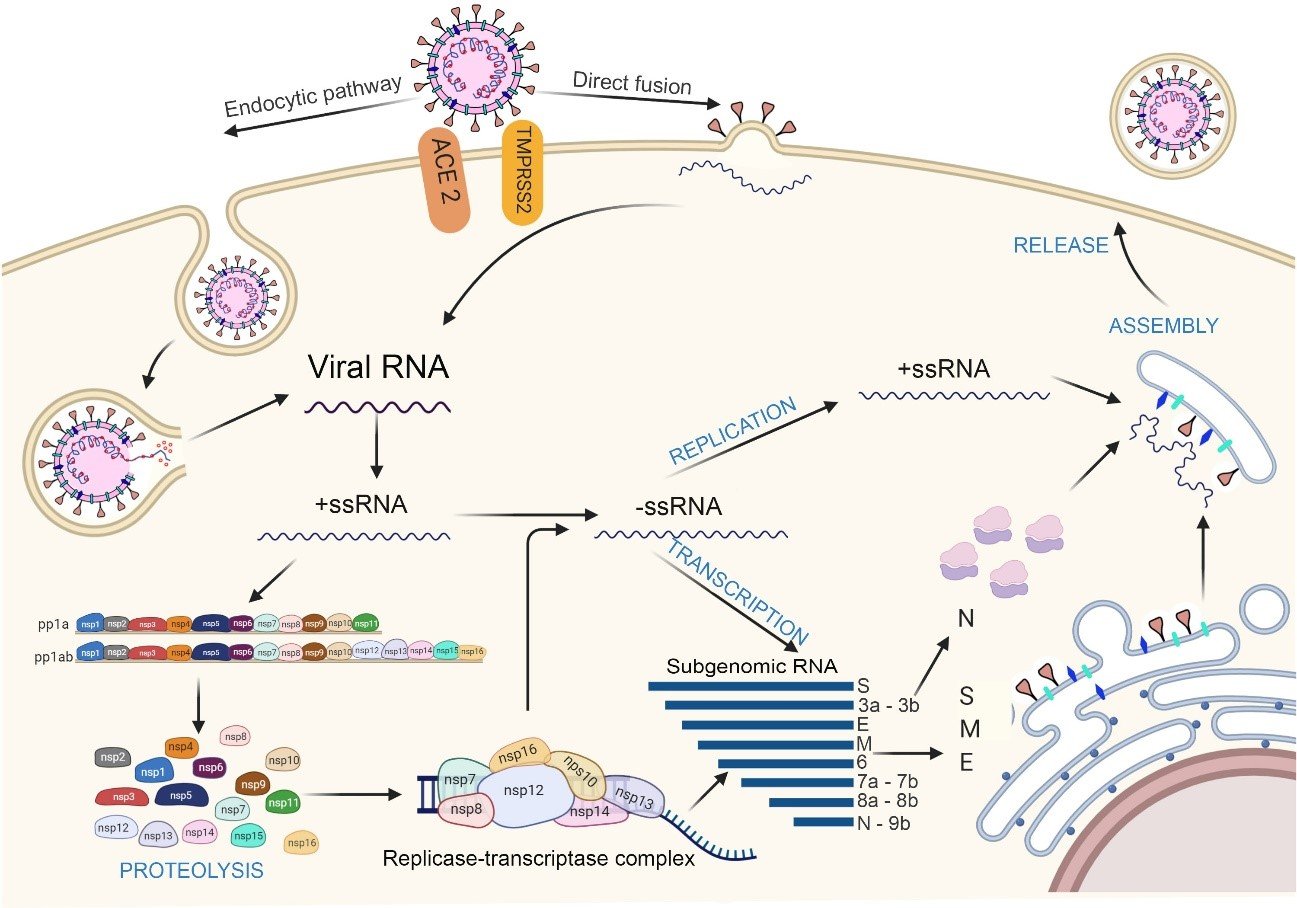
Figure 5. Mechanism of interaction and pathogenesis of SARS-CoV-2. Entry of SARS-CoV-2 through 2 pathways: endocytosis (to the left) and direct fusion (to the right). Genomic RNA enters the cell and positive single-stranded RNA (mc+RNA) is translated into the polyproteins pp1a and pp1ab from the ORF 1a and ORF 1ab regions. Subsequently, a process of autoclivation by 3CLpro and Mpro, will give rise to the 16 non-structural proteins (Nsps), which will form the replicate transcriptase complex (RTC), which will produce single-stranded negative polarity RNA from the positive strand; which will be associated with the nucleocapsid protein. On the other hand, the RTC complex, will synthesize subgenomic RNA (sg RNA) that will encode the S, M and E proteins, assembled in endoplasmic reticulum before being transported to the RE-Golgi compartment, where it will associate with the new genomic RNA and the N protein. Finally, it will be exported in the form of vesicles for the subsequent release of the new virus. Elaborated with https://biorender.com/
SARS-CoV-2 GENOMIC ORGANIZATION
SARS-CoV-2 belongs to the genus of betacoronaviruses and shares a genome sequence identity of 79.6% with SARS-CoV and 50% with MERS-CoV(5). It has a single-stranded positive genomic RNA (ssRNA) with a length of approximately 30 Kb(6); in addition, it has a polyadenylated tail (poly-A) at the 3'-end and a methylated cap at the 5'-end, having a structural similarity to the messenger RNA (mRNA) of eukaryotic cells(15).
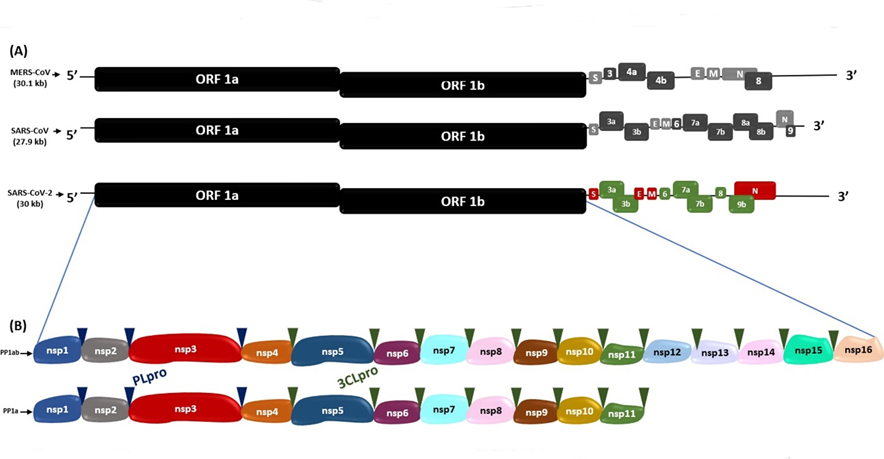
This RNA consists of 15 open reading frames (ORFs) which are RNA sequences comprised between a translation initiation codon and a termination codon(16). In SARS-CoV, MERS-CoV and SARS-CoV-2(16), in the two thirds near the 5'-end of their genome, there are the ORF1a and ORF1b which encode the polyproteins 1a (PP1a) and 1ab (PP1ab), respectively; the cleavage of these polyproteins originates the non-structural proteins (Nsp1-16), which make up the viral replicase-transcriptase complex(16). On the other hand, in the third part near the 3’-end, there are the ORFs S, E, M and N, which encode the spike (S), envelope (E), nucleocapsid (N) and membrane (M) proteins, respectively, and the ORFs 3a, 3b, 6, 7a, 7b, 8, 9a, 9b and 10 that encode the accessory proteins(16,17) (Figure 1).
Figure 1. Genomic sequences of SARS-CoV-2, MERS-CoV and SARS-CoV (A) Genomic sequence comparison of SARS-CoV-2 with MERS-CoV and SARS-CoV. (B) Polyprotein 1ab (PP1ab) and polyprotein 1a (PP1a) encoded by ORF 1a and ORF 1b, with their respective non-structural proteins (Nsps). The blue and green triangles indicate the cleavage sites of the PLpro and 3CLpro proteases, respectively.
SARS-CoV-2 PROTEINS
The knowledge of proteins and their functions in SARS-CoV-2 has been obtained mainly from several researches on SARS-CoV and other related betacoronaviruses (such as MERS-CoV), but in less proportion, due to the similarities of the SARS-CoV-2 genome with these group of viruses(5,18). Table 1
shows the percentages of identity of SARS-CoV-2 proteins with their SARS-CoV counterparts(6,16,19-24).
SARS-CoV-2 has 16 non-structural proteins, four structural proteins and eight accessory proteins(16) , which will be described below.
Table 1. Porcentaje de identidad entre proteínas de SARS-CoV-2 y SARS-CoV.
|
Protein |
% Identity with SARS-CoV |
|
Nsp1 |
86% |
|
Nsp2 |
<70% |
|
Nsp3 |
76% |
|
Nsp4 |
80% |
|
Nsp5 |
97% |
|
Nsp6 |
88% |
|
Nsp7 |
67% |
|
Nsp8 |
85% |
|
Nsp9 |
97% |
|
Nsp10 |
99% |
|
Nsp12 |
97% |
|
Nsp13 |
100% |
|
Nsp14 |
95% |
|
Nsp15 |
88% |
|
Nsp16 |
94% |
|
Spike |
75% |
|
Envelope |
89% |
|
Membrane |
90.50% |
|
Nucleocapsid |
90.52% |
Non-Structural Proteins
The 16 non-structural proteins come from the proteolytic cleavage of the polyproteins PP1a and PP1ab expressed by ORF1a and ORF1ab(16), respectively. They play a fundamental role in the replication of the virus inside the host cells and some of them are the target of several drugs in development(7,25).
Nsp1
Its structure has a 6-stranded beta barrel with an alpha helix covering a terminal part of the barrel and another along it(26).
This protein interacts with the ribosomal 40S subunit of the host cell by through the residues Lys164 and His165, using this subunit to get access to the host mRNA(27). The Nsp1 does not allow the binding of the 40S ribosomal subunit to the 60S subunit, thus inhibiting mRNA translation(27). It then recruits an endonuclease that induces endonucleotic cleavage at the 5'-UTR of the mRNA; this leads to degradation of the 5’-truncated intermediate by the exoribonuclease Xrn1(27).
Nsp2
It presents a structure stabilized by Gln321, due to the length of its side chain, polarity and potential to form hydrogen bonds(28).
It binds to the host cell's PHB 1 and PHB2 (prohibitin 1 and 2) proteins, which are involved in cell cycle progression, cell migration, cell differentiation, apoptosis and mitochondrial biogenesis(28). However, the mechanism of Nsp2 binding to PHB1 and PHB2 proteins has not been studied yet(18).
Nsp3
It is the largest SARS-CoV-2 protein and from the N-terminus, it sequentially presents an ubiquitin-like domain that binds to single-stranded RNA, an ADP-ribose-binding module, a single-stranded poly(A) binding domain, a papain-like viral protease, a nucleic acid-binding domain and a G protein-coupled C receptor(29).
This protein, together with Nsp4 and Nsp6, regulates the replication site by recruiting the replicating protein to the host membrane(28). The multi-layer transmembrane domains of Nsp3 protein serve as a scaffold for the assembly of the replicase-transcriptase complex associated with the membrane(30).
The papain-like protease (PLpro) domain of Nsp3 (Figure 2) is responsible for the release of Nsp1, Nsp2 and Nsp3 from the N-terminus of the polyproteins 1a and 1ab(29). Also, this domain can bind to RIG-I, NEMO, TRAF6 proteins so that they do not activate the transcription factors IRF3 and NFKB, which coordinate the expression of type I interferons; because of this, the production of important cytokines involved in the activation of the host's innate immune response against viral infection is blocked(29,31).
Nsp4
This transmembrane protein presents multiple substitutions near its N-terminus and has a quite preserved C-terminus; these regions are cytosolic(18). Besides, it has a very conserved domain similar to human defensins involved in innate immunity which is constituted by amino acid residues that extend from positions 217 to 237(32).
The coexpression of Nsp4 with the C-terminal one-third of Nsp3 occurs in positions 112-164 in its luminal loop, thus allowing the redistribution of the endoplasmic reticulum to the perinuclear region for the induction of double membrane vesicle formation, but when expressed individually Nsp4 is located in the endoplasmic reticulum(33). Also, when expressed together with Nsp6, they allow optimal replication within the host cells. On the other hand, it has been observed that in the amino acid residues His120 and Phe121 in SARS-CoV, Nsp4 plays a crucial role in membrane remodeling byits interaction with Nsp3(33).
Nsp5
The SARS-CoV-2 main protease (Mpro, NSP5, 3CLpro) is a highly conserved 67.6 kDa homodimeric cysteine protease that differs by only 12 amino acids with the corresponding M pro protease of SARS-CoV(20).
It consists of domains I (residues 8-101), II (residues 102-184) and III (residues 201-303), with a long loop (residues 185-200) connecting domains II and III(20) (Figure 2). The Glu166 residue is a key amino acid involved in the dimerization of Mpro and in the creation of substrate binding pockets(34). In addition, Cys141 and His41 residues form a catalytic dyad at the active site of the protein, essential for its function(20,34,35).
This protease is cleaved from the polyproteins to produce mature enzymes, and then cleaves more downstream non-structural proteins at 11 sites to release Nsp4 - Nsp16. In addition, it acts as a mediator in the maturation of Nsps, which is essential in the life cycle of the virus(34,36).
Nsp6
It is a transmembrane protein consisting of 290 amino acids and located in the endoplasmic reticulum(37). The part of its structure located in the region of the external membrane has multiple phenylalanine residues, which facilitates the affinity of this protein with the membrane of the reticulum and would make its binding more stable(38).
Nsp6 forms complexes with Nsp3 and Nsp4; in addition, it is involved in the formation of double membrane vesicles derived from endoplasmic reticulum during coronavirus replication(21,37). It is hypothesized that the formation of these double membrane vesicles are induced as omegasomes by the inhibition of mammalian target of rapamycin (mTOR) by Nsp6 or by the activation of autophagy through the induction of alternate pathways, which continue until the formation of the autophagosome which in normal conditions it would form an autolysosome to degrade its content, in this case viral, by the infection of SARS-CoV-2(21,37). However, this process would not take place thanks to Nsp6 that would form smaller autophagosomes (less than 0.5um) than normal (approx. 1um). It is suggested that the reduced size of these autophagosomes limits their ability to fuse with lysosomes, this would benefit viral replication by preventing maturation of endosomal and autophage vesicles, consequently, their ability to degrade viral elements, and providing them with new machinery for replication under safe conditions(21,37).
Nsp7 y Nsp8
The structure of Nsp7 and Nsp8 are predominantly alpha-helix(38,39) (Figure 3). Both form a hexadecameric supercomplex that adopts a hollow cylindrical structure and can participate in viral replication by acting as a primase, whose positive electrostatic properties imply that it confers procesivity to RNA-dependent polymerase(40).
Nsp9
It has a core made up of a small seven-stranded enclosed β barrel, from which a series of extended loops projected outwards(22). The elongated loops join the individual β strands of the barrel, together with a projection N-terminal β strand and C-terminal α1 helix; these last two elements constitute the main components of the dimeric arrangement of the protein(22,41). There are two ways for the dimerization of Nsp9, one is the interaction that occurs between the parallel α-helices of each monomer that contain the protein-protein interaction motif Gli-X-X-Gli interaction motif, and the other way is through a beta-sheet interface stabilized by main chain atom interactions within the sheet regions of each monomer(41).
The Nsp9 has the ability to bind RNA and DNA, thus mediating viral replication, general virulence and viral reproduction of genomic RNA; it is probably a member of the replication complex(22).
Nsp10
The structure of this small protein of 139 amino acids (Figure 3) is formed by a pair of antiparallel beta-sheet in the center, surrounded on one side by a large loop that crosses them and by 5 alpha-helices whose loops form two zinc finger domains, the first zinc binding site is coordinated by the Cys74, Cys77, His83 and Cys90 residues and the second zinc binding site is coordinated by Cys117, Cys120, Cys128 and Cys130(16, 42). The function of these sites in other coronaviruses is involved with non-specific binding to RNA(42).
The C-terminal domain of Nsp10 forms multiple interactions with the ExoN domain of Nsp14, which strongly affects its nucleolytic activity, which improves up to 35 times(43) . On the other hand, this protein also acts as a cofactor of Nsp16, increasing the activity of its 2'-O-MTase domain(43).
Nsp12
In its structure we find a N-terminal β hairpin in the residues 31-50, a NiRAN (nidovirus RdRp-associated nucleotidyl-transferase) domain in the residues 115-250 with seven helices and three β strands, an interface domain in the 251-365 residues composed of three helices and five β strands that connects the NiRAN domain and the RdRp domain, which has a hollowed-out configuration, with fingers subdomains in the 397-581 and 621-679 residues, a thumb subdomain in the 819-920 residues, and a palm subdomain, which form a closed circle(44) (Figure 3).
The RdRp domain has RNA dependent polymerase activity, but by itself it presents low activity; therefore, it requires accessory factors, which are the Nsp7 and Nsp8 proteins with which it forms a complex, which presents motifs with conserved Zn2+ binding residues in Cys487 His642, Cys645, Cys646 and in His295, Cys301, Cys306, Cys310(44). This increases the bonding of RdRp to the RNA mold-first(40,44).
Nsp13
Its structure has five domains: ZBD, stalk, 1b, 1a and 2a domains; and an active NTPasa site which is composed of Lys288, Ser289, Asp374, Glu375, Gln404 and Arg567; these are vital for the unwinding of a duplex RNA molecule(45) (Figura 3).
At high ATP concentrations, the protein's helicase activity has an increased affinity for duplex RNA, which develops in three steps: First, Nsp13 binds to the 5′-ss tail in the presence of ATP without ATP hydrolysis; then, by adding magnesium ions it triggers ATP hydrolysis and finally, Nsp13 allows the separation of the duplex RNA and translocates along the unwound RNA in the 5' to 3' direction(46).
Nsp14
This protein has an N-terminal domain (ExoN) that includes three motifs (I(DE), II(E), III(D)) and a C-terminal domain containing (guanine-N7)-methyl transferase (N7-MTase)(47). The ExoN domain has an alpha/beta fold, composed of a central beta-sheet formed by five beta-strands flanked by alpha-helices, with the exception of beta-3, and its catalytic residues include Asp90, Glu92 Glu191, Asp272(47). On the other hand, the N7-MTape domain is formed by a beta-sheet consisting of five beta-strands and a canonical S-adenosylmethionine (SAM)(47) binding motif. A hinge region separates the ExoN domain from the N7-MTase domain, this is flexible and consists of a loop and three strands, allowing lateral and rotational movements of the two domains to coordinate enzymatic activities(43,47).
The Nsp14 function as a methyltransferase dependent on S-adenosyl methionine (SAM) (guanine-N7) (N7-MTase)(47). After hydrolysis of the nascent RNA by Nsp13, p-RNA is formed. An unknown guanylyltransferase (GTase) hydrolyzes GTP, transfers the GMP product to pRNA and creates Gpp-RNA. Then, SAM methylates the guanine 5' of the Gppp-RNA at the N7 position, forming the 5'(m7GpppN- RNA) cap(43,47).
Nsp15
This protein is a specific nidoviral RNA uridylate endoribonuclease (NendoU) with monomeric units composed of 345 amino acids that fold into three domains(48). The N-terminal domain is composed of an antiparallel β-sheet wrapped around two α-helices (α1 and α2) and the subsequent middle domain is formed by 10 β-strands organized in three pins, a mixed β-sheet, and three short helices(48). The NendoU catalytic domain of the C terminal contains two antiparallel β-sheets with edges that host a catalytic site(48).
The active site is located in a not too deep groove between the two β-sheets, it carries six key residues: His235, His250, Lys290, Thr341, Tyr343 and Ser294, of which, the residues His235, His250 and Lys290 constitute the catalytic triad, while Ser294 and Tyr343 are in charge of the NendoU specificity(48,49).
Nsp16
Its structure contains a highly conserved catalytic tetrad (Lys-Asp-Lys-Glu), distinctive of the RNA 2′-O-MTas, inside a nucleus composed of a Rossmann-type β-sheet fold decorated by eleven α helix, seven β-strands and loops(50). In addition, it forms the SARS-CoV-2 fold, formed by a β-sheet which is encased by loops and α propellers(50) (Figure 3).
Structural proteins
Four structural proteins have been identified in SARS-CoV-2, which are also present in other coronaviruses. These are the Spike(S), Envelope(E), Membrane(M) and Nucleocapsid(N) proteins (Figure 4).
Spike(S) protein
This protein has a molecular weight of 180 kDa(16). Its structure contains the functional subunits S1 and S2, located in its ectodomain(51).
The S1 subunit has an N-terminal domain, a C-terminal domain(51) and a conserved receptor binding domain (RBD) containing a core and a receptor binding motif (RBM)(52). This subunit mediates the binding to the ACE 2 receptor, where amino acid residues such as Lys317 and Phe486 from the RBD domain could be key to this interaction(51,52).
On the other hand, the S2 subunit has in its structure a fusion peptide domain (FP), an heptad-1 and -2 repetition domains (HR1, HR2) and a transmembrane domain (TM), which allow the fusion of the viral and cellular membranes(51,52).
S protein requires a protease cleavage for the activation of its fusion potential. Two sequential steps have been proposed for the cleavage model; an initial cleavage between S1 and S2, and the subsequent cleavage activation at the S2' site(53).
In addition, it presents a large mutated surface, with four new inserts in the protein, three of them are located in the first NTD domain, while the fourth is located immediately before the cleavage S2 site and within the homo-trimerization interaction interface(16). Likewise, the RBD domain is not affected by these inserts, but it is the most mutated region with potential alterations in its function of binding to ACE 2(16).
Envelope Protein (E)
It has many similarities with the sequences of other coronaviruses; however, there are distinctive characteristics such as the substitution of glutamate, glutamine or aspartate residues by arginine in the position and the replacement of the Ser-Phe dyad by Thr-Val in positions 55-56(54).
This protein is the smallest of the four structural proteins, with 76 amino acids in length(55,56). Its structure has a negatively charged short amino-hydrophilic end consisting of 7 to 12 amino acids, followed by a large 25-amino-acid hydrophobic transmembrane (TMD) domain, and ends with a variable-load long hydrophilic carboxyl end (55,56). The hydrophobic region of the TMD contains an amphipatic α helix that oligomerizes to form an ion-conducting pore in the membranes, a part of the TMD consists of two non-polar neutral amino acids, Val and Leu, which confer a strong hydrophobicity to the protein(56) . The C-terminal end also exhibits some hydrophobicity, but less than the TMD due to the presence of a positively charged basic amino acid group, and also contains a preserved proline residue centered on a β-coil-β motif, which probably functions as a direction signal to the Golgi complex(56).
Membrane Protein(M)
This integral membrane glycoprotein is the more abundant of the four proteins and provides the morphology of the virion(57,58). It has a length of approximately 220-260 amino acids with a short length N-terminal domain, integrated into the virus membrane through three transmembrane domains labeled as tm1, tm2 and tm3(57-59) . Its glycosylated short amino terminal end constitutes an ectodomain outside the membrane, while its C-terminal endodomain is located on the cytoplasmic side of the virion membrane(57,58). The ectodomain can be glycosylated, affecting the tropism of the organs to be infected and the interferon-inducing capacity (IFN) of some coronaviruses(58,59). Also, it presents the insertion of a serine residue in position 4 as a unique feature in SARS-CoV-2(23).
During assembly, it provides a scaffold for the viral particles, stabilizes the N protein (N - RNA protein complex) and the inner core of the virions; it is also necessary for the retention of the S protein in the ER-Golgi intermediate compartment (ERGIC) and its incorporation to new virions(57). The coexpression of M and E form the viral envelope, their interaction is sufficient for the production and release of virus-like particles (VLP)(58,59).
Nucleocapsid Protein (N)
Its structure is made of two well-folded domains, known as the N-terminal domain (NTD) and the C-terminal domain (CTD)(24). Both domains are rich in β-strands, but CTD also has some short helices(24).
It binds directly to the viral RNA and provides stability to it(60). In addition, it has been found to antagonize antiviral RNAi and inhibit the activity of the cyclin-CDK (cyclin-cyclin-dependent kinase) complex; this inactivation results in the hydrophosphorylation of the retinoblastoma protein and in turn inhibits the progression of the S phase in the cell cycle(18).
Accessory proteins
SARS-CoV-2 accessory proteins are expressed by the genes ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9a, ORF9b and ORF10. Several of these proteins have functions still unknown and it is suspected that they do not intervene in viral replication but may have important roles in viral pathogenesis(18).
PATHOGENESIS OF COVID-19
Virus entry and binding to ACE2 receptor
SARS-CoV-2 can enter the host cell by two routes: endocytosis or direct fusion with the cell surface(15,51). For the endocytosis route, the virus is encapsulated by the endosome after binding to its ACE2 receptor(15). Then, the low pH environment promotes cleavage of the S protein with the pH-dependent L cysteine protease (CPL)(50). On the other hand, in the direct fusion route with the cell surface, after the binding of the RBD domain of the S1 subunit to the ACE2 receptor, the transmembrane protease serine 2 (TMPRSS2) splits and activates the S protein, in the ectodomain formed by S1 and S2, which allows the fusion of the viral membrane with the host cell membrane(15,51,61).
The activation of the S-protein is performed in the cleavage site S1/S2 also called "polybasic site" or "multibasic site" which contains several arginine residues (Ser-Pro-Arg-Ala-Arg ↓ Ser)(15,61). A second cut is also needed at S2′ in Lys-Arg ↓ Ser-Phe(53), which is no different from other SARS-CoV-2-like viruses; it activates the fusion peptide (FP) that binds the host membranes and the virus, constituting the intermediate stage of fusion(53,62,63). Finally, the segment between HR1 and HR2 changes its conformation, they bind, forming a heptamer (6-HB) that joins both membranes facilitating the entry of the virus(61).
It is also known that other proprotein-converting enzymes, such as furan or trypsin from the host cell, recognize the cleavage S1/S2 site(61).
SARS-CoV-2 replication, transcription and translation mechanism
After the entry of the virus into the host cell, through the endocytic route, endosomal acidification is required, that is to say a decrease in pH mediated by lysosomes(64), to unite the endosome membrane with the viral envelope, releasing its nucleocapsid to the cytoplasm(64,65).
The RNA virus in the ORF1a and ORF1b regions acts as mRNA where the viral replicase gene is directly transcribed with the host cell machinery, translating into the polyproteins PP1a and PP1ab(15). These are split by the papain like proteases (PLpro; corresponding to Nsp3) and 3C like (3CLpro or Mpro; corresponding to Nsp5) to form the 16 non-structural proteins (Nsps)(15,64).
The nonstructural proteins are then reorganized into double-membrane vesicles from the endoplasmic reticulum (ER)(64), then assembled in the perinuclear region, in the replica-transcriptase complex (RTC), creating a suitable environment for the synthesis of negative (-) RNA through replication and transcription, so that it replicates and synthesizes a set of subgenomic mRNAs (sgRNA)(64,65).
Subgenomic RNAs are synthesized by combining variable lengths of the 3' end of the genome with the 5' leader sequence required for translation. These subgenomic (-) RNAs are transcribed into subgenomic (+) mRNAs, which encode the structural proteins S, M, E, N and the accessory proteins (towards the 3' end)(64,65).
The newly synthesized viral genomic RNA binds to the N protein originating the nucleocapsid(64). S, M, E and accessory proteins, expressed from sgRNAs, are synthesized in the ER membranes and then transported to the Golgi complex to be assembled with the nucleocapsid, producing new viral particles, which through the vesicle transport system, will travel to the surface and will be release by exocytosis(64,65) (Figure 5).
As previously mentioned, SARS-CoV-2 uses ACE2 as receptor, which is expressed in different cells of the vascular system, central nervous system, eyes, upper airways, heart, lungs and intestine; being the last three more vulnerable than the others(66) (66) (Figure 6).
IN SILICO STUDIES OF POTENTIAL PHARMACOLOGICAL TREATMENT
The expression in silico is used to characterize those studies that are completely executed using computer models.(67) It means that the environment and behavior of the object of study under this method are represented by models that simulate its relevant characteristics through software engineering.(67) The advantage of these studies is the speed of execution, low cost and the ability to reduce the use of animals; therefore, their use has been a strategy to accelerate the discovery of potential new drugs such as those described in this section(67). The design of drug prototypes in silico, in the studies cited in this article, mainly covers the structure-activity relationship, in this case between SARS-CoV-2 proteins and drug molecules(67).
Proteins as functional units of the cell represent a major site of pharmacological and xenobiotic action(68). Therefore, characterizing the target proteins is fundamental to understand the mechanism of action of drugs and xenobiotics(68). The coupling of substrates at the active site of a known or modeled protein structure can be used to explain protein-substrate interactions and possibly predict the chemicals that will interact with the proteins(69).
Understanding how different amino acids interact with substrates (ligand, protein, drug, etc.) is crucial to explain the conformation and affinity of protein-ligand interactions(68). This technique, also known as structure-based drug design, uses existing 3D protein structure data to predict protein-ligand interactions using computer-aided ligand docking software(69). Docking uses an energy-based scoring function, with lower energy scores representing better protein-ligand bonds compared to higher energy values(70). The scoring function is defined by the sum of the energy of ligand-protein interaction and the internal energy of the ligand(70).
The molecules that have been found in many in silico studies have been divided into two groups: the ones approved for human use that are intended to be repurposed to treat SARS-CoV-2(71-73) (Table 1) and the plant origin ones(74-76) (Table 2).
Table 2. Drugs approved for human use that are intended to be reproposed to treat SARS-CoV-2
|
Author, Year, Reference number |
Molecule |
Expected effect on SARS-CoV-2 structure |
Expected result |
Common use in medicine |
|
Sarma, P; et al. 2020 [71] |
8- (2-hydroxyethyl) aminophylline |
Binding to the N-terminal domain of the N protein |
Replication inhibition |
Bronchodilator |
|
Sarma, P; et al. 2020 [71] |
ethyl (4S) -4-methyl-2-oxo-6 - [(1S) -1-phenylethyl] -3,4-dihydro-1H-pyrimidine-5-carboxylate |
Binding to the N-terminal domain of the N protein |
Replication inhibition |
Antiviral |
|
Lobo-Galo, N; et al. 2020 [72] |
Disulfiram |
Viral protease inhibition |
Replication inhibition |
Alcoholism |
|
Mahanta, S; et al. 2020 [73] |
Viomycin |
Attack on the active site of the Mpro |
Replication inhibition |
Tuberculosis |
CONCLUSIONs
The Nsps participate in the viral cycle of the virus and favor its infection; while the structural proteins mediate the union and fusion of the virus to the host cell, as well as its assembly; until now the functions of the accessory proteins are unknown. The activation of the S protein by TMPRSS2, furin, trypsin or catepsins allows the entry of SARS-CoV-2 into the host cell; where its viral RNA translates the proteins of the replicase-transcriptase complex, which initiates the synthesis of a template for new genomic RNAs, which translate the structural proteins. In silico studies of many reproposed and plant origin molecules have shown their inhibitory action on SARS-CoV-2 proteins Nsp3, Nsp5, N, M and E.
ACKNOWLEDGEMENTS
The molecular graphics in Figures 2 and 3 were made using UCSF Chimera by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Table 3. Molecules of natural origin suitable for human use for treatment against COVID-19.
|
Author, Year, Reference number |
Molecule |
Expected effect on SARS-CoV-2 structure |
Result |
Source |
|
Borkotoky, S; et al. 2020 [74] |
Nimbolin A |
Inhibition of ionic channels of E protein and interacts mainly with the transmembrane domain of M protein |
Prevents the spread of the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
Nimocin |
Inhibition of ionic channels of E protein and interacts mainly with the transmembrane domain of M protein |
They can make it difficult to assemble the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
7-Deacetyl-7-Benzoylgedunin |
Inhibition of ionic channels of E protein |
Prevents the spread of the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
24-Methylenecycloartanol |
Inhibition of ionic channels of E protein |
Prevents the spread of the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
Cycloeucalenone |
Inhibition of ionic channels of E protein |
Prevents the spread of the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
Phytosterol |
Interact with the C-terminal region, mainly with the conserved CD domain of the M protein |
They can make it difficult to assemble the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
Beta-Amyrin |
Interact with the C-terminal region, mainly with the conserved CD domain of the M proteins |
They can make it difficult to assemble the virus |
Azadirachta indica (Neem) |
|
Borkotoky, S; et al. 2020 [74] |
24-Methylenecycloartan-3-one |
Interact with the C-terminal region, mainly with the conserved CD domain of the M protein |
They can make it difficult to assemble the virus |
Azadirachta indica (Neem) |
|
Bhardwaj, VK; et al. 2020 [75] |
Oolonghomobisflavan-A |
Interact with the active site of Mpro by binding to critical waste. |
Replication inhibition |
Planta de té |
|
Bhardwaj, VK; et al. 2020 [75] |
Theasinensin-D |
Interact with the active site of Mpro by joining to critical waste. |
Replication inhibition |
Planta de té |
|
Bhardwaj, VK; et al. 2020 [75] |
Theaflavin-3-O-gallate |
Interact with the active site of the Mpro by joining to critical waste |
Replication inhibition |
Planta de té |
|
Enmozhi, SK; et al. 2020 [76] |
Andrographolide |
Interact in the binding site of Mpro |
Replication inhibition |
Andrographis paniculata |
Authorship contributions: The authors participated in the genesis of the idea, project design, data collection and interpretation, analysis of results and preparation of the manuscript of this research work.
Funding sources: Self-funded.
Conflict of interest: The authors declare no conflict of interest.
Received: October 14, 2020.
Approved: January 07, 2021.
Correspondence: Eduardo Rubén Lam Cabanillas.
Address: Prol. Costa Rica Mz R’ Lote 7 Dpto. 102, Urb. Monserrate Trujillo-Perú.
Telephone: 953 015 443
E-mail: elam@unitru.edu.pe
REFERENCIAS BIBLIOGRAFICAS