Introduction
Bacterial resistance to antimicrobials has a genetic basis that can be intrinsic and/or extrinsic to the microorganism. In recent years, it has gained importance due to the increase in in-hospital mortality, largely because of the irrational use of antimicrobials, which has led it to be declared one of the top 10 threats to public health
1
. Additionally, healthcare costs have risen due to the use of expensive antibiotics, longer hospital stays, and a social impact on the wages and productivity of the affected patient
2
.
It is known that the spectrum of antimicrobial resistance is variable, thus microorganisms are classified as multidrug-resistant (MDR) when they show no sensitivity to at least one agent in three or more antimicrobial categories; extensively drug-resistant (XDR) when they show no sensitivity to at least one agent in all categories except one or two, that is, bacterial isolates remain sensitive to only one or two families; and pandrug-resistant (PDR) microorganisms when they show no sensitivity to any agents in all categories of antimicrobials, meaning that no tested agents are effective for that organism
3
.
The microbiological map is a document that provides information about the microorganisms circulating in different hospital departments, evaluates the sensitivity of these microorganisms to the antimicrobials in use, thus contributing to the initiation of an effective and timely empirical treatment in patients with infections, reducing hospital stay, and decreasing healthcare costs
4
.
In some hospitals in Peru, such as in Lima, Arequipa, or Trujillo, microbiological maps have been developed, but in many other regions of the country, this information is lacking. It is important to mention that after the COVID-19 pandemic, the irrational use of antimicrobials for the treatment of SARS-CoV-2 pneumonia significantly contributed to the advance of antimicrobial resistance
5
. Due to the lack of a local microbiological map and the high number of reported infections by multidrug-resistant pathogens, this study aims to identify the microorganisms and their antimicrobial resistance profiles in infections developed in clinical and surgical services.
Methodology
This is a descriptive cross-sectional study that evaluated the results of cultures and antibiograms from clinical samples of patients with suspected infection, who were admitted between January and December 2021 in a hospital located in the Peruvian Jungle. This hospital is one of the main healthcare centers in the Loreto region, with 191 hospital beds, including 56 in the Emergency Department: 42 for adults, 10 for pediatrics, and 4 for obstetrics and gynecology. Additionally, there are 21 beds in the Intensive Care Unit (ICU): 11 for adults, 4 for neonates, and 6 for pediatrics. Other departments include 14 beds in Pediatrics, 26 in Surgery, 19 in Obstetrics and Gynecology, 14 in Traumatology, 29 in Internal Medicine, and 12 in Infectious Diseases. The hospital serves a population of 240,000 people.
For microbiological identification, the VITEK® 2 (bioMérieux) system was used, which is an automated spectrophotometric system for assessing microbial (bacteria and fungi) growth and susceptibility to antimicrobials, according to updated cutoff points from the Clinical and Laboratory Standards Institute (CLSI) guidelines. The type of microorganism, antimicrobial sensitivity, source of isolation, patient's age, and hospital department were identified.
The study was approved by the hospital's Ethics Committee. Data were collected in an Excel spreadsheet (version 2016), coded, tabulated, and analyzed using SPSS Statistics version 25.00. Descriptive statistical analysis was performed, including measures of central tendency and frequency distribution.
Results
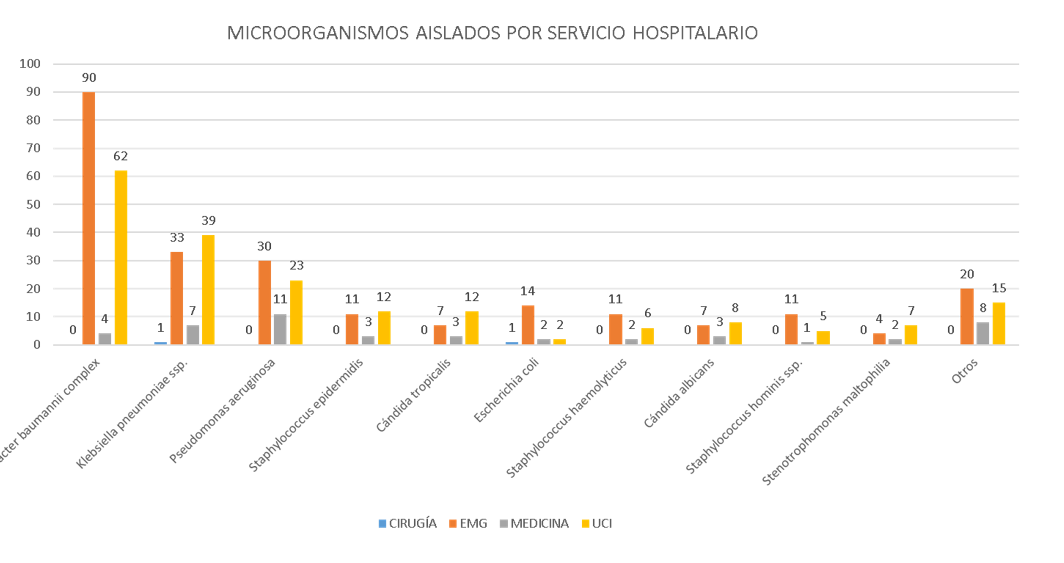
A total of 477 positive cultures were identified from 453 patients; 262 (54.9%) cultures came from bronchial secretion samples, 168 (35.2%) from blood cultures, 31 (6.5%) from urine cultures, and five or fewer from wound secretion, laminar drain, central venous catheter tip, tracheal secretion, cerebrospinal fluid, pleural fluid, and peritoneal secretion samples. The age of the patients ranged from one day old to 88 years (median of 56 years). The microorganisms most regularly isolated were *Acinetobacter baumannii* complex and *Klebsiella pneumoniae* ssp., found most frequently in bronchial secretion cultures from the Emergency Department and Intensive Care Units (Figure 1).
Among the group of Gram-negative bacteria, *Acinetobacter baumannii* complex showed 53% sensitivity to tigecycline and 97% to colistin, while it exhibited 97% resistance to meropenem and 98% to imipenem, being XDR in 88.5% of cases. Regarding *Klebsiella pneumoniae* ssp., 58% were ESBL (+), with 10% sensitivity to ceftazidime and 100% resistance to ceftriaxone, in addition to 44% and 34% resistance to meropenem and imipenem, respectively, being MDR in 56.3% of cases. In the case of *Pseudomonas aeruginosa*, it showed 96% sensitivity to colistin and 70% to amikacin, along with 85% and 82% resistance to meropenem and imipenem, being XDR in 54.7% of cases. As for *Escherichia coli*, 89% were ESBL (+) with 6% sensitivity to ceftazidime and 13% to ceftriaxone (Table 1).
Regarding Gram-positive bacteria, all were isolated from blood cultures; *Staphylococcus epidermidis* was the most frequent, followed by *Staphylococcus haemolyticus*, with 88% and 100% methicillin resistance, respectively, and no resistance to vancomycin was reported for either microorganism. As for *Staphylococcus aureus*, 67% were methicillin-resistant, with no reported vancomycin resistance. Regarding *Enterococcus faecium*, the only isolated strain was resistant to ampicillin but sensitive to vancomycin (Table 2).
In terms of fungi, they were isolated in a total of 43 samples, of which the antifungal susceptibility profile was performed in only six cultures; *Candida parapsilosis* was found in the blood culture of a 66-year-old patient, showing resistance to fluconazole and voriconazole (Table 3).
Figura 1.
Microorganismo aislado según servicio hospitalario en un nosocomio general de la selva peruana 2021
EMG: Emergencias, UCI: Unidad de Cuidados Intensivos Adulto + Unidad de Cuidados Intensivos Neonatales. Otros: Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae complex, Morganella morganii ssp., Proteus hauseri, Proteus mirabilis, Providencia stuartii, Pseudomonas stutzeri, Serratia marcescens, Sphingomonas paucimobilis, Enterococcus faecalis, Enterococcus faecium, Kocuria kristinae, Staphylococcus aureus, Staphylococcus capitis, Staphylococcus lugdunensis, Staphylococcus saprophyticus, Staphylococcus warneri, Cándida ciferrii, Cándida krusei, Cándida parapsilosis.